PROSUP+
PROSUP+ stands for the prosupination movement of the distal radioulnar joint (DRUJ). The goal of this project is to study the morphology and biomechanical factors around the DRUJ for simulating not only the healthy but also the pathological forearm motion with respect to functional disability and instability. The ultimate goal of the project is to be able to predict the surgical outcome preoperatively by analyzing the forearm motion before and after the simulated surgery.
PROSUP+ is a multi-center project fully funded by the Swiss National Science Foundation (SNF) and the Austrian Research Fund (FWF). It focuses on the analysis and simulation of the distal forearm stability during pro-supination for improved surgical planning.
The distal radioulnar joint (DRUJ) is one of two joints of the forearm bones radius and ulna. It is anatomically located at the wrist. The articulating bone surfaces enable forearm motion (i.e., pro-supination) in an extensive range while the stability is primarily maintained by a complex system of ligaments connecting the bones. Consequently, posttraumatic soft tissue injury can cause chronic joint instability, pain, or functional disability. In these patients, operative treatment by anatomical reconstruction of the injured ligament using a tendon graft is indicated. Bone realignment by corrective osteotomy is additionally mandatory if ligament tear is accompanied by posttraumatic bone deformity. The surgical outcome, however, remains unpredictable in many cases because the effect of the surgery on the forearm mobility cannot be accurately assessed preoperatively. The reason for this is that state-of-the-art computer simulations do not take into account any soft tissue structures. For corrective surgeries of the forearm bones, computer assisted planning relies only on the comparison to the healthy bone of the opposite side. These approaches can obviously not be applied to pathologies where soft tissue components play a key role. Methods are therefore needed to allow the patient-specific modelling and subsequent simulation of soft tissue structures of the forearm during pro-supination motion.
The project is divided into three main parts, namely: (1) anatomical modelling, (2) simulation, and (3) clinical validation. The key idea for understanding and learning the motion pattern is first the acquisition of static and dynamic data of the forearm through ex-vivo experiments using fresh-frozen cadaver forearms. The modelling part will focus on the development of methods for generating a 3-dimensional representation of the relevant patient anatomy (i.e., bones with ligaments) from computed tomography and magnetic resonance images in an automatic fashion. The data obtained from the cadaver experiments will serve as a basis for the development of the computer model of the forearm motion. The simulation model will be embedded into an existing preoperative planning framework for predicting functional improvement. Lastly, the methods will be validated against the cadaver experiments.
Generation of Bone- and Soft-Tissue 3D Models: Interosseous Membrane
The interosseous membrane (IOM) is the biggest ligamentous structure in the forearm, connecting the radius and ulna bones in the diaphysis. The kinematics of the pro-supination and the range of motion reached by a patient is strongly influenced by the IOM. Previous studies have demonstrated that the IOM highly influences bone motion and forearm stability in healthy and pathological cases. Both, in-vivo and cadaveric studies have shown that the motion of the forearm cannot be considered isolated from the IOM and that structures inside the IOM have a major role in the elasticity of the entire ligament. In practice, it is not uncommon to observe cases of forearm surgeries where the final outcome was unexpected due to the influence of the IOM. These findings make clear the need of a forearm simulation models capable of including the influence of soft tissue in order to improve the reliability and accuracy of preoperative planning.
For the simulation model of the IOM, several anatomical and biomechanical parameters are needed. In order to generate 3D morphological accurate IOM models, the thicknesses, widths and attachments of the ligament structures and their corresponding insertion angles along the forearm axis are needed. Secondly, for modelling the biomechanical behavior, the elastic properties of each ligament structure is required, namely, the tensile and shear modulus, the cross-sectional area (CSA), elastic and shear stiffness and maximum tensile and shear forces.
Morphological properties corresponding to each of the individual ligaments of the IOM have been only reported by Noda et al. (2009) using simple 2D measurement techniques and without reporting elastic properties. Several cadaveric studies have focused on the description of the biomechanical properties of the IOM, but the reported results are not sufficient to build a complete 3D model of the ligaments.
In this project part, new 3D measurement methods have been developed for the generation of precise morphological data of the interosseous membrane. In a first step, highly precise 3D models of the IOM were generated from cadaveric forearms (Figure 1) through a combination of micro- and standard-CT imaging. Afterwards, our 3D measurement methods were applied to these 3D models in order to evaluate morphological features of the IOM (Figure 2). Morphological values were validated in cadaver experiments against established, optical measurement methods (Figure 3) and available literature. Additionally, the individual tensile properties of each IOM ligament were investigated in biomechanical experiments.
The generated 3D models have been made publicly available in other to contribute towards the improvement of existing forearm models.
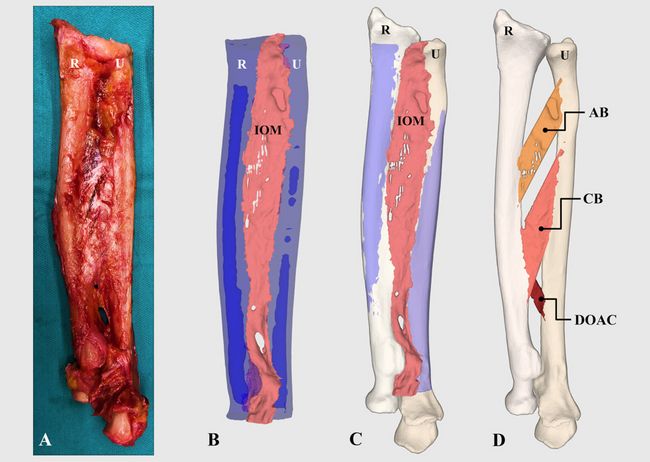
Figure 1: Generation of 3D models of forearm ligaments. (A) IOM after resection of all muscles. (B) Model of the IOM (shown in pink), radius (R) and ulna (U) (shown in blue) segmented from the micro-CT data. (C) The entire forearm model is build by aligning the micro-CT-based 3D models to a standard-CT-based model of the entire forearm. (D) 3D models of the IOM ligaments.
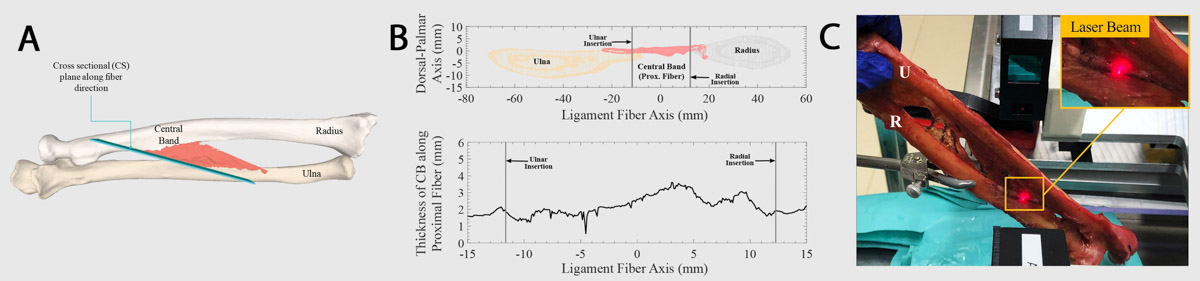
Figure 2: Thickness measurement for the 3D models of the IOM ligaments. (A) Cross-sectional plane defined in the generated 3D models of the IOM (B) Cross-sectional cut of the radius, ulna and IOM surface meshes, generated by a plane aligned at the proximal fiber of the central band (CB) ligament of the IOM and thickness values for the CB calculated from the cross-sectional cut along the proximal fiber.
3D-reconstruction of soft- and hard-tissues through image fusion
This part of the project addresses the problem of data fusion from different modalities with the goal of 3D-reconstructing a combined soft- and hard-tissue 3D model of the forearm. The approach is based on a 3D image-to-image registration pipeline consisting of 3 main steps: A manual coarse alignment of the image data (in our case MRI and CT), an automatic identification of the region of interest in order to avoid anatomical misalignment, and an automatic registration based on probabilistic process maximizing the similarity between both datasets using entropy. The method was evaluated on a retrospective series of distal radius cases. A detailed description of the method can be found in Gerber et al3.
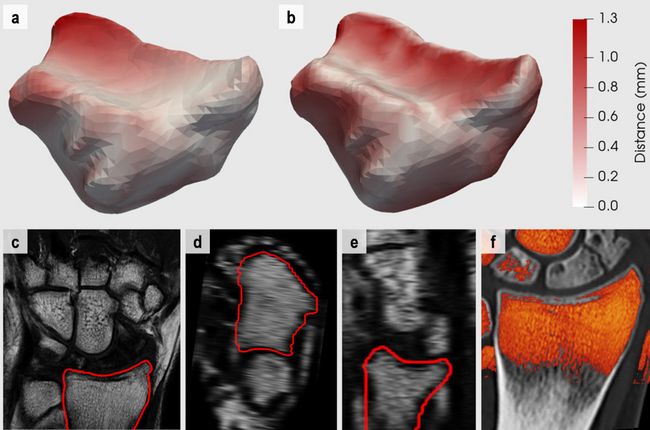
Figure 4: Results representation of CT to MR registration of the distal radius. a-b) Colour-encoded Euclidean error distances mapped onto the 3D surface of the distal radius extracted from the CT data for the a) proposed automatic pipeline and the b) landmark-based registration methodology. White to red colour gradient maps to the absolute error distances [0.0 – 1.3] millimeters. c-e) MR image data in c) coronal, d) axial and e) sagittal planes including the outline of the 3D radius surface extracted from the aligned CT. f) Colour-encoded overlay of the MR image data onto the CT data of the distal radius. Red to yellow colours map to the intensity value range of [0 – 270] on our data sets in order to include soft tissue while excluding cortical bone.
Forearm Motion from Cadaveric Data
We are currently developing a motion model of the forearm based on ground truth data obtained from human cadaveric samples. A custom-made and motor-driven device has been constructed in which the cadavers can be placed to simulate the forearm pro-supination motion (Figure 5A). The goal of this project part is to study the influence of each of the ligament structures on the stability and motion pattern of the forearm. The motion is acquired using a commercial motion tracking system (Figure 5B).
Simulation Pilot
A computer simulation of the pro-supination motion, which considers the influence of the IOM, has been performed together with the Computer Vision Lab (CVL) of ETH Zürich (Pèan et al., 2017). Ligament attachments and elastic behaviour were extracted from previous anatomical studies. The collision of ligaments was also implemented in order to enable wrapping around the bones. The outcome of the simulation was compared to available data from the literature.
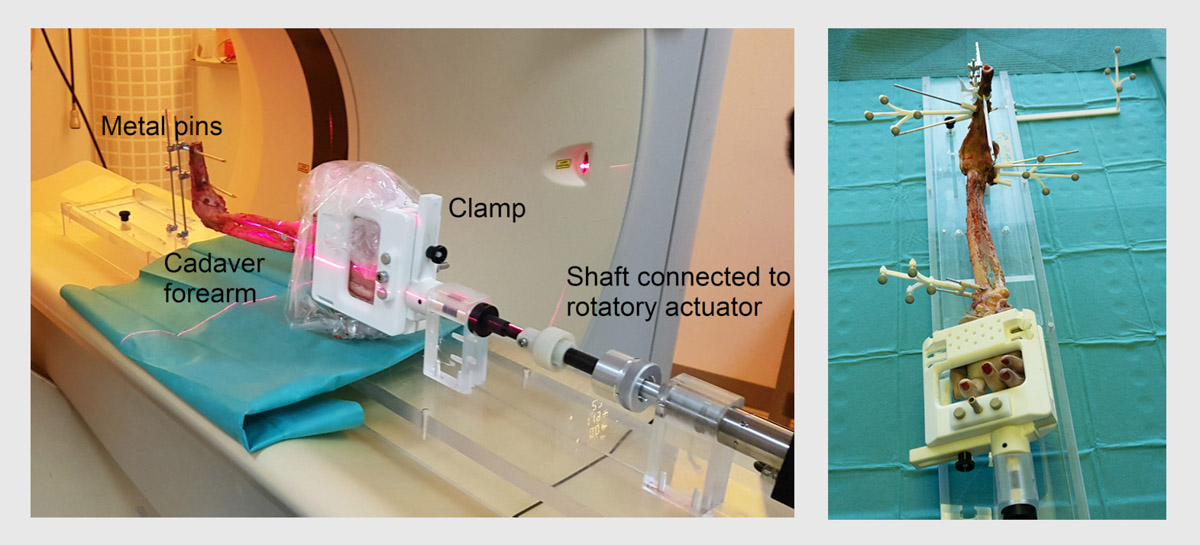
Figure 5. (A) Setup of the pro-supination device for motion simulation on the cadaver. (B) Tracking system for motion analysis of the forearm.
References
1. Carrillo, F., Gerber, N., Abegg, D., Sutter, R., Nagy, L., Zheng, G., Fürnstahl, P. Automated CT-MR image fusion for the preoperative planning of orthopedic surgeries, Swiss Orthopaedics Jahreskongress (SGOT) 2019.
2. Péan, F., Carrillo, F., Fürnstahl, P., Goksel, O., 2017. Physical Simulation of the Interosseous Ligaments During Forearm Rotation, In: Klaus Radermacher, F.R.Y.B. (Ed.), CAOS 2017. 17th Annual Meeting of the International Society for Computer Assisted Orthopaedic Surgery, pp. 181--188.
3. Gerber N., Carrillo F., Abegg D., Sutter R., Zheng G., Fürnstahl P. Evaluation of CT - MR Image Registration Methodologies for 3D Preoperative Planning of Forearm Surgeries. Journal of Orthopaedic Research, February 2020.